Research Topics
Nucleic Acid Sensors in Innate Immunity
We are interested in how the innate immune system first detects evading pathogens and the mechanisms in place to prevent autoimmunity. The innate immune system is a phylogenetically ancient defense mechanism that is present in all multicellular organisms and has evolved a limited set of nonclonal, germ-line encoded pathogen recognition receptors (PRRs) to sense evading pathogens. Each of these receptors is specialized in detecting one type of pathogen-associated (PAMP) or damage-associated molecular pattern (DAMP). Nucleic acids, such as DNA or RNA, which are used by any organism to carry genetic information, can serve as PAMPs and are one of the most important classes of molecules recognized by our immune system. However, nucleic acids are not limited to pathogens, but also present within the host. This blurs the lines between self and non-self-discrimination and puts host nucleic acids at risk of being recognized by innate immune sensors, which can lead to autoimmune and autoinflammatory diseases. Here we aim to understand how these immune receptors recognize specific nucleic acids together with their sophisticated strategies to overcome self-recognition. From an evolutionary perspective, we are interested in how these immune receptors have adapted to recognize a particular ligand and what signaling mechanisms are present to fine-tune the immune response across evolution to match the species-specific pathogen burden.
Our group focuses on a recently discovered new family of nucleic acid sensing proteins including the DNA sensing pathway cGAS–STING as well as the RNA sensing pathway OAS1–RNase L. Upon sensing pathogenic double-stranded DNA, cyclic GMP–AMP synthase (cGAS) catalyzes the production of cyclic 2′–5′ GMP 3′–5′ AMP (2′3′-cGAMP) from common nucleotide triphosphate precursors ATP and GTP. 2′3′-cGAMP is recognized by the adaptor protein Stimulator of Interferon Genes (STING) and triggers a signaling cascade leading to the production of type I interferons and pro-inflammatory cytokines. Oas proteins recognize dsRNA instead and upon recognition can produce the linear second messenger 2'-5'-oligo adenylate (2'-5'OA) of varying length from ATP. This linear nucleotide second messenger can then activate the downstream restriction factor RNase L responsible for cellular as well as viral RNA cleavage leading to the inhibition of protein synthesis and apoptosis limiting viral spread.
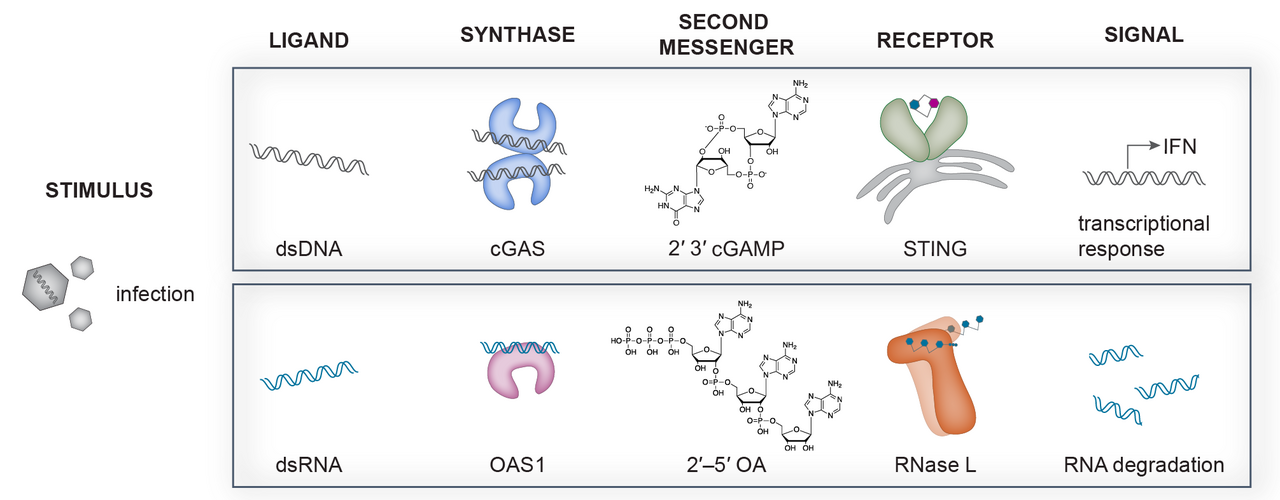
Nucleic Acid Sensing NTase pathways in Innate Immunity
Nucleotidyltransferases (NTases) and Nucleotide-based Second Messenger Signaling
We aim to reveal the molecular function of several uncharacterized human NTases and discover novel nucleotide-based second messengers in humans. cGAS and OAS proteins belong to the large and highly diverse superfamily of nucleotidyltransferases (NTases). NTases share a common structural fold besides low sequence homology and require a ligand induced conformational switch for activation. Their common catalytic mechanism involves the transfer of nucleoside monophosphate (NMP) to a variety of substrates, including RNA/DNA, proteins, sugars, NTPs, and small molecules, with the list of potential substrates and products growing with each newly characterized NTase reaction mechanism. Several subfamilies of uncharacterized human NTases and their products remain to be discovered, despite their importance in biological processes ranging from immunity to development. Consistent with this, patient mutations in NTase family members result in drastic phenotypes and are associated to rare genetic diseases as well as cancer. Our work will be critical in revealing the molecular mechanism causing these diseases and developing novel therapeutic strategies using nucleotide-based second messengers as drugs. A challenge to reveal the molecular function of these NTases is the requirement of a yet unknown ligand for activation and product synthesis. We apply protein-nucleic acid biochemistry, structural biology using x-ray crystallography and single particle cryo-electron microscopy combined with cellular assays and small molecule mass spectrometry to identify and study NTase ligands and their products.
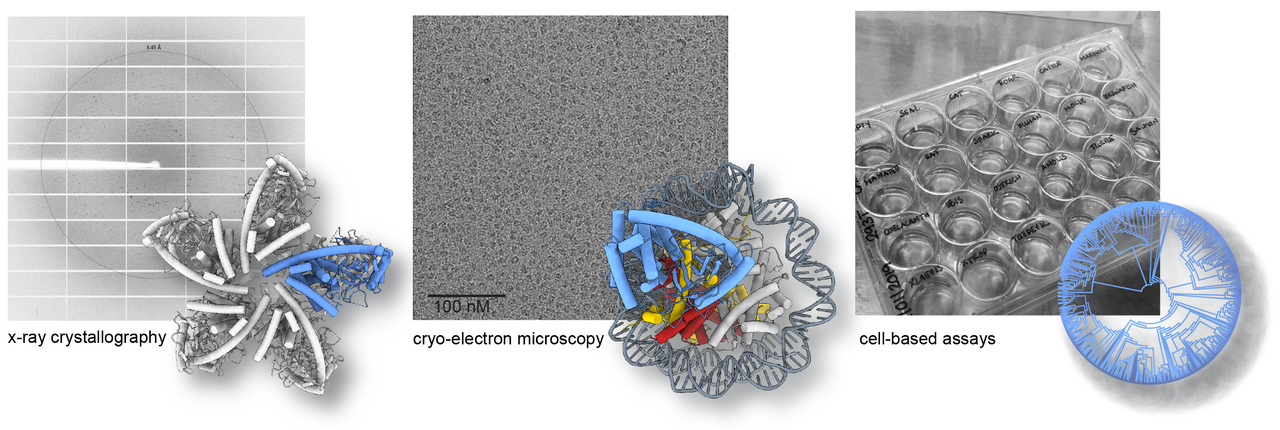
Methods to study NTases and their activation mechanisms